CLI Submissions
Introduction
This guide is for anyone seeking guidance on submitting data to an Overture platform. By the end of this guide you will have completed a full data submission workflow, including updating the submitted data to conform to the data model enforced by Song.
You will need docker installed. We recommend using Docker Desktop; for more information, visit Dockers website
Background: Submitting data to an Overture platform typically includes data files (BAMS, CRAMS, VCFs, etc.) along with associated metadata that provides context including donor information and descriptions of the data files. This guide focuses on submitting data to Song & Score using their Command Line Clients (CLIs).
The Song and Score clients work together to upload raw data files to object storage and track all related metadata stored in the Song metadata repository.
Getting Started
1. Clone the Quickstart repository
git clone -b quickstart https://github.com/overture-stack/prelude.git
2. Launch the platform by running the appropriate command for your operating system:
- For Unix/macOS run
make platform - For Windows run
./make.bat platform
Ensure enough resources get allocated to Docker We recommend a minimum CPU limit of 8, memory limit of 8 GB, swap of 2 GB, and virtual disk limit of 64 GB. You can access these settings by selecting the cog wheel found on the top right of the Docker desktop app and selecting resources from the left panel. If you already have docker desktop installed be ensure you are on version 4.32.0 or higher
CLI Submssion
Step 1: Generate an API Key
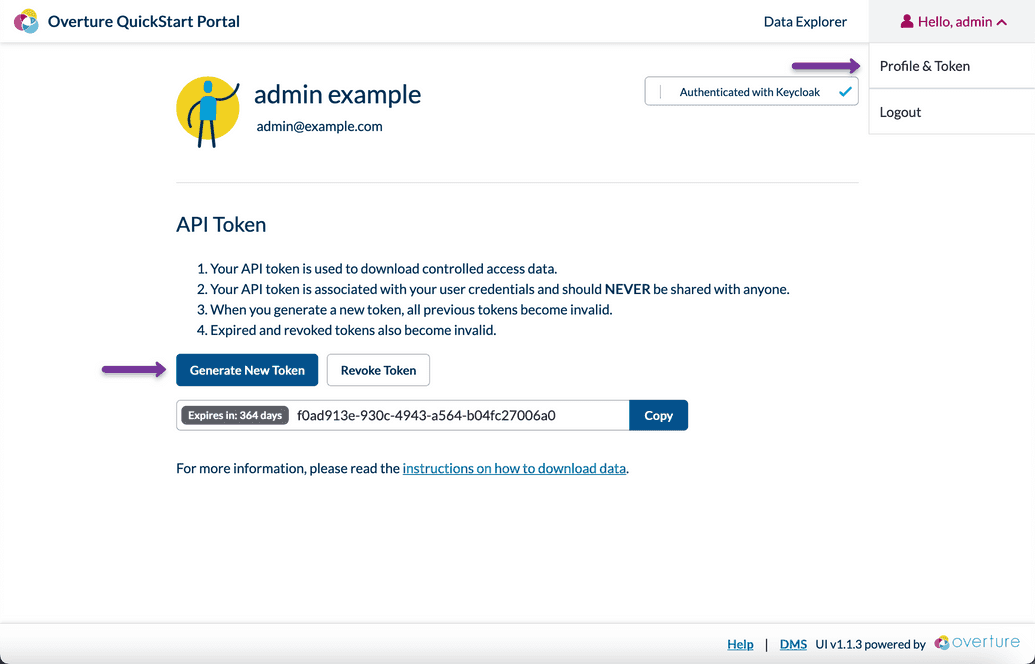
API Keys are brokered by Keycloak and accessible when logged in to the Stage UI. For the Overture QuickStart, Stage can access from localhost:3000
Login through the Stage UI by selecting login from the top right. The default credentials will be username
adminand passwordadmin123when using the Overture QuickStart.Once logged in, it's time to generate a new API token: To do this, click on Profile and Token from your user drop down menu, located on the top right of the Stage UI. Then, select Generate New Token.
Step 2: Run the Song and Score Clients
- Running the Song Client: Use the following command with your API token to pull and run a Song Client docker container
docker run -d -it --name song-client \-e CLIENT_ACCESS_TOKEN=68fb42b4-f1ed-4e8c-beab-3724b99fe528 \-e CLIENT_STUDY_ID=demo \-e CLIENT_SERVER_URL=http://localhost:8080 \--network="host" \--platform="linux/amd64" \--mount type=bind,source=./guideMaterials/dataSubmission,target=/output \ghcr.io/overture-stack/song-client:5.1.1
Click here for a detailed breakdown
-druns the container in detached mode, meaning it runs in the background and does not receive input or display output in the terminal-itcombines the-i(interactive) and-t(allocate a pseudo-TTY) options, allowing you to interact with the container via the terminal-e CLIENT_ACCESS_TOKEN=68fb42b4-f1ed-4e8c-beab-3724b99fe528sets up the song-client with a pre-configured system-wide access token. Alternatively, you can log in through Stage fromlocalhost:3000/loginwith the usernameadminand passwordadmin123. From the profile page, you can generate your API key and supply it here-e CLIENT_STUDY_ID=demothe quickstart is pre-configured with aStudy IDnameddemo, we supply theStudy IDvalue to the song-client on start-up-e CLIENT_SERVER_URL=http://localhost:8080is the url for the Song server which the Song-Client will interact with--network="host"Uses the host network stack inside the container, bypassing the usual network isolation. This means the container shares the network namespace with the host machine--platform="linux/amd64"Specifies the platform the container should emulate. In this case, it's set to linux/amd64, indicating the container is intended to run on a Linux system with an AMD64 architecture--mount type=bind,source=./guideMaterials/dataSubmission,target=/outputmounts the directory and its contents (volume) from the host machine to the container. In this case, the mockData being used for our submission. It binds the directory ./guideMaterials/dataSubmission from the host to /output inside the container. Any changes made to the files in this directory will be reflected in both locations
Note: Ensure you are running the following commands from the root directory of the quickstart folder. The values here reflect those compatible with the Overture QuickStart.
- Running the Score Client: Use the following command with your API token to pull and run a Score Client docker container
docker run -d -it --name score-client \-e ACCESSTOKEN=68fb42b4-f1ed-4e8c-beab-3724b99fe528 \-e STORAGE_URL=http://localhost:8087 \-e METADATA_URL=http://localhost:8080 \--network="host" \--platform="linux/amd64" \--mount type=bind,source=./guideMaterials/dataSubmission,target=/output \ghcr.io/overture-stack/score:latest
Click here for a detailed breakdown
-druns the container in detached mode, meaning it runs in the background and does not receive input or display output in the terminal-itcombines the-i(interactive) and-t(allocate a pseudo-TTY) options, allowing you to interact with the container via the terminal-e ACCESSTOKEN=68fb42b4-f1ed-4e8c-beab-3724b99fe528sets up the score-client with a pre-configured system-wide access token. Alternatively, you can log in to Stage fromlocalhost:3000/loginwith the usernameadminand passwordadmin123. From the profile page, you can generate your API key and supply it here-e STORAGE_URL=http://score:8087is the url for the Score server that the Score-Client will interact with-e METADATA_URL=http://song:8080is the url for the song server that the score-client will interact with--network="host"Uses the host network stack inside the container, bypassing the usual network isolation. This means the container shares the network namespace with the host machine--platform="linux/amd64"Specifies the platform the container should emulate. In this case, it's set to linux/amd64, indicating the container is intended to run on a Linux system with an AMD64 architecture--mount type=bind,source=./guideMaterials/dataSubmission,target=/outputmounts the directory and its contents (volume) from the host machine to the container. In this case, the mockData being used for our submission. It binds the directory ./guideMaterials/dataSubmission from the host to /output inside the container. Any changes made to the files in this directory will be reflected in both locations
Note: Ensure you are running the following commands from the root directory of the quickstart folder. The values here reflect those compatible with theclea Overture QuickStart.
Step 3: Submit metadata to Song
What are we submitting?
We will now begin submitting our payload to the Overture platform. In this context a payload refers to a collection of related metadata and file data to be uploaded to the resource.
The mock data we will use can be found in the Overture Quickstart repository from the quickstart/guideMaterials/dataSubmission/ directory
The files included are two VCF data files (
SP059902.snv.vcf.gzandSP059902.snv.vcf.gz.tbi) and a JSON file,SP059902.vcf.jsonthat contains all the associated metadata
Click here to view the metadata file (SP059902.vcf.json)
{"studyId": "demo","analysisType": {"name": "quickStartSchema"},"collaborator": [{"name": "IICR","contactEmail": "dataSubmitter@example.com"}],"samples": [{"submitterSampleId": "SP059902","sampleType": "Total DNA","matchedNormalSubmitterSampleId": null,"specimen": {"submitterSpecimenId": "SP059902","specimenType": "Normal - tissue adjacent to primary tumour","tumourNormalDesignation": "Normal","specimenTissueSource": "Blood derived - bone marrow"},"donor": {"submitterDonorId": "DO0599","gender": "Female"}}],"files": [{"dataType": "Raw SV Calls","fileName": "SP059902.snv.vcf.gz","fileSize": 17246,"fileMd5sum": "94b790078d8e98ad08ffc42389e2fa68","fileAccess": "open","fileType": "VCF","info": {"dataCategory": "Simple Nucelotide Variation","jbrowseCoordinates": "hg38:chr1:100000-200000"}},{"dataType": "Raw SV Calls","fileName": "SP059902.snv.vcf.gz.tbi","fileSize": 141,"fileMd5sum": "f5cca6ace25d076d1f76cebf4ce3defd","fileAccess": "open","fileType": "TBI","info": {"dataCategory": "Simple Nucelotide Variation","jbrowseCoordinates": "hg38:chr1:100000-200000"}}],"specimen": {"submitterPrimaryDiagnosisId": "PD059901","submitterSpecimenId": "SP059902","specimenAnatomicLocation": "C31","tumourGradingSystem": "Nuclear grading system for DCIS","tumourGrade": "G1"},"donor": {"submitterDonorId": "DO0599","primarySite": "Trachea","vitalStatus": "Alive","survivalTime": null,"causeOfDeath": null,"primaryDiagnosis": [{"submitterPrimaryDiagnosisId": "PD059901","ageAtDiagnosis": 50,"cancerTypeCode": "C34.2","clinicalTumourStagingSystem": "Binet staging system","clinicalStageGroup": "Stage A","treatment": [{"submitterTreatmentId": "TR059901","treatmentType": ["Chemotherapy"],"treatmentStartInterval": 58,"treatmentDuration": 52,"responseToTreatment": "Complete response","chemotherapy": [{"drugName": "Tamoxifen "}]}],"followUp": [{"submitterFollowUpId": "FO059901","submitterTreatmentId": "TR059901","intervalOfFollowUp": 45,"diseaseStatusAtFollowUp": "Stable","relapseType": null}]}]},"experiment": {"platform": "PacBio","experimentalStrategy": "WXS","model": "SEQUEL IIe","sequencingCenter": "CGTA","sequencingDate": "2021-03-08T19:00:00.000Z"},"workflow": {"workflowName": "Mutect2 Variant Calling","workflowShortName": "Mutect2Variant","workflowVersion": "0.1.1.1","genomeBuild": "GRCh38_hla_decoy_ebv","inputs": [{"analysisType": "sequencing_alignment","tumourAnalysisId": "00000000-0000-0000-0000-0000000000599","normalAnalysisId": "00000000-0000-0000-0000-0000000000599"}],"runId": "RI0599","sessionId": "SI0599"},"publication": {"publication": "NAR","doi": "10.1093/nar/gkae188"}}
The Submission Command
First we will submit the JSON metadta file to Song using the song-client submit command:
docker exec song-client sh -c "sing submit -f /output/SP059902.vcf.json"
Expected Error Response
When submitting data, Song checks the metadata against the specified data model, identified by the analysisType field located on line 4 of the SP059902.vcf.json file. The error logs indicate that the submitted metadata does not conform to this data model:
SONG_SERVER_ERROR[schema.violation @ 1719691299284]: [SubmitService::schema.violation] - #/collaborator/0: required key [name] not found,#/donor/primarySite: Windpipe is not a valid enum value
The error log tells us the following:
We are missing the required
collaborator.namefieldThe value
Windpipeis not valid entry for the fieldprimarySite
Addressing the missing field:
We will address our first error by updating our metadata payload with a collaborator name. To do this we will append lines 6 to 10 as follows:
"collaborator": [{"name": "IICR","contactEmail": "dataSubmitter@example.com"}],
Feel free to provide any name value you like, in this example we are submitting from the Imaginary Instituite for Cancer Researcher (IIRC).
Addressing the invalid value:
To address our second error we will have to take a look at the resources data model. For convenience we are providing the relevant Song schema below however, if you're interested in learning how to access this information through Songs API, we've provided additional instructions below.
View Song Schema
{"name": "quickStartSchema","schema": {"type": "object","required": ["donor", "specimen", "workflow", "experiment"],"properties": {"workflow": {"propertyNames": {"enum": ["workflowName","workflowShortName","workflowVersion","genomeBuild","inputs","sessionId","runId"]},"required": ["workflowName", "genomeBuild", "inputs"],"type": "object","properties": {"workflowName": {"type": "string","pattern": "^[a-zA-Z][a-zA-Z0-9 _\\-]+[a-zA-Z0-9]+$"},"workflowShortName": {"type": "string","pattern": "^[a-zA-Z][a-zA-Z0-9_\\-]+[a-zA-Z0-9]+$"},"workflowVersion": {"type": "string"},"genomeBuild": {"type": "string","enum": ["GRCh37", "GRCh38_hla_decoy_ebv", "GRCh38_Verily_v1"]},"inputs": {"type": "array","items": {"type": "object","properties": {"tumourAnalysisId": {"type": "string","pattern": "^[0-9a-f]{8}-[0-9a-f]{4}-[0-9a-f]{4}-[0-9a-f]{4}-[0-9a-f]{13}$"},"normalAnalysisId": {"type": "string","pattern": "^[0-9a-f]{8}-[0-9a-f]{4}-[0-9a-f]{4}-[0-9a-f]{4}-[0-9a-f]{13}$"},"analysisType": {"type": "string"}}},"minItems": 1,"maxItems": 2},"runId": {"type": "string"},"sessionId": {"type": "string","pattern": "SI[0-9]{4}"}}},"experiment": {"propertyNames": {"enum": ["platform","experimentalStrategy","model","sequencingCenter","sequencingDate"]},"required": ["platform", "experimentalStrategy"],"type": "object","properties": {"platform": {"type": ["string", "null"]},"experimentalStrategy": {"type": ["string", "null"]},"model": {"type": ["string", "null"]},"sequencingCenter": {"type": ["string", "null"]},"sequencingDate": {"type": ["string", "null"],"pattern": "^(-?(?:[1-9][0-9]*)?[0-9]{4})-(1[0-2]|0[1-9])-(3[01]|0[1-9]|[12][0-9])T(2[0-3]|[01][0-9]):([0-5][0-9]):([0-5][0-9])(.[0-9]+)?(Z|[+-](?:2[0-3]|[01][0-9]):[0-5][0-9])?$"}}},"donor": {"type": "object","propertyNames": {"enum": ["submitterDonorId","primarySite","vitalStatus","survivalTime","causeOfDeath","primaryDiagnosis"]},"required": ["submitterDonorId","primarySite","vitalStatus","primaryDiagnosis"],"properties": {"submitterDonorId": {"type": "string"},"primarySite": {"type": "string","enum": ["Accessory sinuses","Adrenal gland","Base of tongue","Bladder","Bones, joints and articular cartilage of limbs","Bones, joints and articular cartilage of other and unspecified sites","Brain","Breast","Bronchus and lung","Cervix uteri","Colon","Connective, subcutaneous and other soft tissues","Corpus uteri","Esophagus","Eye and adnexa","Floor of mouth","Gallbladder","Gum","Heart, mediastinum, and pleura","Hematopoietic and reticuloendothelial systems","Hypopharynx","Kidney","Larynx","Lip","Liver and intrahepatic bile ducts","Lymph nodes","Meninges","Nasal cavity and middle ear","Nasopharynx","Oropharynx","Other and ill-defined digestive organs","Other and ill-defined sites","Other and ill-defined sites in lip, oral cavity and pharynx","Other and ill-defined sites within respiratory system and intrathoracic organs","Other and unspecified female genital organs","Other and unspecified major salivary glands","Other and unspecified male genital organs","Other and unspecified parts of biliary tract","Other and unspecified parts of mouth","Other and unspecified parts of tongue","Other and unspecified urinary organs","Other endocrine glands and related structures","Ovary","Palate","Pancreas","Parotid gland","Penis","Peripheral nerves and autonomic nervous system","Placenta","Prostate gland","Pyriform sinus","Rectosigmoid junction","Rectum","Renal pelvis","Retroperitoneum and peritoneum","Skin","Small intestine","Spinal cord, cranial nerves, and other parts of central nervous system","Stomach","Testis","Thymus","Thyroid gland","Tonsil","Trachea","Ureter","Uterus, NOS","Vagina","Vulva"]},"vitalStatus": {"type": "string","enum": ["Alive", "Deceased"]},"survivalTime": {"type": ["null", "integer"],"minimum": 0},"causeOfDeath": {"type": ["null", "string"],"enum": ["Died of cancer", "Died of other reasons", "Unknown", null]},"primaryDiagnosis": {"type": "array","minItems": 1,"items": {"type": "object","propertyNames": {"enum": ["submitterPrimaryDiagnosisId","ageAtDiagnosis","cancerTypeCode","clinicalTumourStagingSystem","clinicalStageGroup","treatment","followUp"]},"required": ["submitterPrimaryDiagnosisId","ageAtDiagnosis","cancerTypeCode"],"properties": {"submitterPrimaryDiagnosisId": {"type": "string"},"ageAtDiagnosis": {"type": "integer","minimum": 0},"cancerTypeCode": {"type": "string","pattern": "^[C|D][0-9]{2}(.[0-9]{1,3}[A-Z]{0,1})?$"},"clinicalTumourStagingSystem": {"type": "string","enum": ["AJCC 8th edition","AJCC 7th edition","AJCC 6th edition","Ann Arbor staging system","Binet staging system","Durie-Salmon staging system","FIGO staging system","Lugano staging system","Rai staging system","Revised International staging system (RISS)","St Jude staging system"]},"clinicalStageGroup": {"type": "string","enum": ["Occult Carcinoma","Stage 0","Stage 0a","Stage 0is","Stage 1","Stage 1A","Stage 1B","Stage A","Stage B","Stage C","Stage I","Stage IA","Stage IA1","Stage IA2","Stage IA3","Stage IAB","Stage IAE","Stage IAES","Stage IAS","Stage IB","Stage IB1","Stage IB2","Stage IBE","Stage IBES","Stage IBS","Stage IC","Stage IE","Stage IEA","Stage IEB","Stage IES","Stage II","Stage II bulky","Stage IIA","Stage IIA1","Stage IIA2","Stage IIAE","Stage IIAES","Stage IIAS","Stage IIB","Stage IIBE","Stage IIBES","Stage IIBS","Stage IIC","Stage IIE","Stage IIEA","Stage IIEB","Stage IIES","Stage III","Stage IIIA","Stage IIIA1","Stage IIIA2","Stage IIIAE","Stage IIIAES","Stage IIIAS","Stage IIIB","Stage IIIBE","Stage IIIBES","Stage IIIBS","Stage IIIC","Stage IIIC1","Stage IIIC2","Stage IIID","Stage IIIE","Stage IIIES","Stage IIIS","Stage IIS","Stage IS","Stage IV","Stage IVA","Stage IVA1","Stage IVA2","Stage IVAE","Stage IVAES","Stage IVAS","Stage IVB","Stage IVBE","Stage IVBES","Stage IVBS","Stage IVC","Stage IVE","Stage IVES","Stage IVS","Cannot be assessed"]},"followUp": {"type": "array","items": {"type": "object","propertyNames": {"enum": ["submitterFollowUpId","intervalOfFollowUp","diseaseStatusAtFollowUp","relapseType","submitterTreatmentId"]},"required": ["submitterFollowUpId","intervalOfFollowUp","diseaseStatusAtFollowUp"],"properties": {"submitterFollowUpId": {"type": "string"},"submitterTreatmentId": {"type": "string"},"intervalOfFollowUp": {"type": "integer","minimum": 0},"diseaseStatusAtFollowUp": {"type": "string","enum": ["Complete remission","Distant progression","Loco-regional progression","No evidence of disease","Partial remission","Progression NOS","Relapse or recurrence","Stable"]},"relapseType": {"type": ["string", "null"],"enum": ["Distant recurrence/metastasis","Local recurrence","Local recurrence and distant metastasis","Progression (liquid tumours)",null]}}}},"treatment": {"type": "array","items": {"type": "object","propertyNames": {"enum": ["submitterTreatmentId","treatmentType","treatmentStartInterval","treatmentDuration","responseToTreatment","chemotherapy","hormoneTherapy","radiation"]},"required": ["submitterTreatmentId", "treatmentType"],"properties": {"submitterTreatmentId": {"type": "string"},"treatmentType": {"type": "array","items": {"type": "string","enum": ["Ablation","Bone marrow transplant","Chemotherapy","Endoscopic therapy","Hormonal therapy","No treatment","Other targeting molecular therapy","Photodynamic therapy","Radiation therapy","Stem cell transplant","Surgery"]}},"treatmentStartInterval": {"type": "integer","minimum": 0},"treatmentDuration": {"type": "integer","minimum": 0},"responseToTreatment": {"type": "string","enum": ["Complete response","Disease progression","NED","Minor response","Partial response","Stable disease"]},"chemotherapy": {"type": "array","items": {"type": "object","propertyNames": {"enum": ["drugName"]},"required": ["drugName"],"properties": {"drugName": {"type": "string"}}}},"hormoneTherapy": {"type": "array","items": {"type": "object","propertyNames": {"enum": ["drugName"]},"required": ["drugName"],"properties": {"drugName": {"type": "string"}}}},"radiation": {"type": "array","items": {"type": "object","propertyNames": {"enum": ["radiationTherapyModality","anatomicalSiteIrradiated"]},"required": ["radiationTherapyModality","anatomicalSiteIrradiated"],"properties": {"radiationTherapyModality": {"type": "string","enum": ["Electron","Heavy Ions","Photon","Proton"]},"anatomicalSiteIrradiated": {"type": "string","enum": ["Abdomen","Body","Brain","Chest","Head","Liver","Lower Limb","Lung","Neck","Pelvis","Skin","Spine","Thorax","Upper Limb"]}}}}},"allOf": [{"if": {"properties": {"treatmentType": {"contains": {"const": "Chemotherapy"}}}},"then": {"required": ["chemotherapy"]}},{"if": {"properties": {"treatmentType": {"contains": {"const": "Radiation therapy"}}}},"then": {"required": ["radiation"]}},{"if": {"properties": {"treatmentType": {"contains": {"const": "Hormonal therapy"}}}},"then": {"required": ["hormoneTherapy"]}}]}}}}}},"if": {"properties": {"vitalStatus": {"const": "Deceased"}}},"then": {"required": ["causeOfDeath", "survivalTime"]}},"specimen": {"type": "object","propertyNames": {"enum": ["submitterSpecimenId","submitterPrimaryDiagnosisId","specimenAnatomicLocation","tumourGradingSystem","tumourGrade"]},"required": ["submitterSpecimenId","submitterPrimaryDiagnosisId","specimenAnatomicLocation"],"properties": {"submitterSpecimenId": {"type": "string"},"submitterPrimaryDiagnosisId": {"type": "string"},"specimenAnatomicLocation": {"type": "string","pattern": "^[C][0-9]{2}(.[0-9]{1})?$"},"tumourGradingSystem": {"type": "string","enum": ["FNCLCC grading system","Four-tier grading system","Gleason grade group system","Grading system for GISTs","Grading system for GNETs","ISUP grading system","Nuclear grading system for DCIS","Scarff-Bloom-Richardson grading system","Three-tier grading system","Two-tier grading system","WHO grading system for CNS tumours"]},"tumourGrade": {"type": "string","enum": ["Low grade","High grade","GX","G1","G2","G3","G4","Low","High","Grade I","Grade II","Grade III","Grade IV","Grade Group 1","Grade Group 2","Grade Group 3","Grade Group 4","Grade Group 5"]}}},"publication": {"type": "object","propertyNames": {"enum": ["publication", "doi"]},"properties": {"publication": {"type": ["string", "null"]},"doi": {"type": ["string", "null"]}}},"collaborator": {"type": "array","items": {"type": "object","propertyNames": {"enum": ["name", "contactEmail"]},"required": ["name"],"properties": {"name": {"type": "string"},"contactEmail": {"type": ["string", "null"],"pattern": "^\\S+@\\S+\\.\\S+$"}}}}}}}
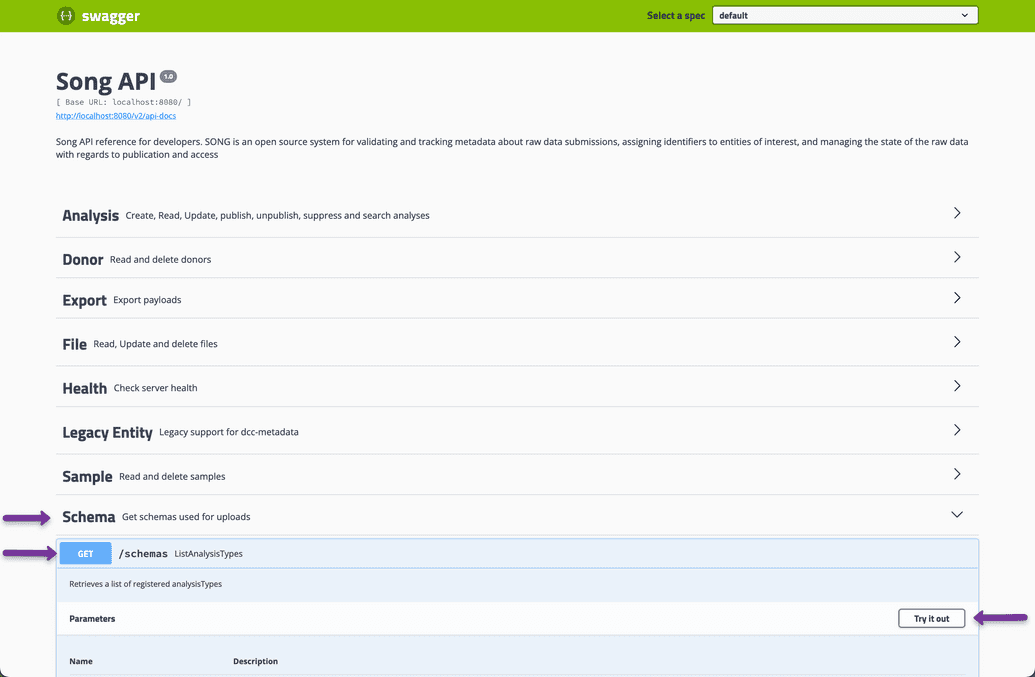
Instructions on accessing the Schema using the Swagger UI click here
- Go to Songs swagger UI located at
localhost:8080/swagger-ui.html - Select Schema and the GET /schemas endpoint
- Click try it out and execute
The response body will provide you a JSON document outlining Songs data model. In this case we are interested in the quickStartSchema as it was the analysisType used for validation of our payload.
By looking at the primarySite field found within the quickStartSchema, we can see an array of values that do not include windpipe; however, it does include the value trachea.
Therefore, we can simply append the windpipe value on line 64 of our SP059902.vcf.json to trachea.
"donor": {"submitterDonorId": "DO0599","primarySite": "Trachea","vitalStatus": "Alive",
Resubmission:
Save and resubmit the corrected metadata file:
docker exec song-client sh -c "sing submit -f /output/SP059902.vcf.json"
Now that your metadata file is correctly formatted, you should recieve a status of OK and a Song generated analysisId
{"analysisId": "4d9ed1c5-1053-4377-9ed1-c51053f3771f","status": "OK"}
An analysis ID is a randomly generated UUID created by Song; as such, yours will differ. Please note your analysis ID and use it for all subsequent steps referencing its usage.
 What is an analysis?
What is an analysis?
Since the metadata data has successfully been submitted and accepted by Song and provided an analysis ID, it is now considered part of a Song analysis. To complete the analysis, we will need to upload its associated file data.
Step 4: Generate a manifest
With your analysis_id, we will now generate a manifest for file upload.
The manifest establishes a link between the analysis ID that has been submitted and the data file(s) on your local system that is being uploaded.
This step also validates that all files being uploaded are in line with those documented in the metadata tagged with the corresponding analysis ID.
Manifest Command
Make sure to replace {AnalysisId} with your previously generated Song analysis ID
docker exec song-client sh -c "sing manifest -a {AnalysisId} -f /output/manifest.txt -d /output/"
The manifest.txt file will be written out to a defined output file path. If the output directory does not exist, it will be automatically created.
Expected Response
Wrote manifest file '/output/manifest.txt' for analysisId '4d9ed1c5-1053-4377-9ed1-c51053f3771f'
Step 5: Upload your data files with Score
Use the following score-client upload command to transfer your file data to object storage using your previously generated manifest.
docker exec score-client sh -c "score-client upload --manifest /output/manifest.txt"
Expected Response
Uploading object: '/output/SP059902.snv.vcf.gz.tbi' using the object id d7442418-7d59-5063-91d4-2e083549c9b2100% [##################################################] Parts: 1/1, Checksum: 100%, Write/sec: 2.0K/s, Read/sec: 0B/sFinalizing...Total execution time: 3.239 sTotal bytes read : 0Total bytes written : 141Upload completedUploading object: '/output/SP059902.snv.vcf.gz' using the object id 0e3ed34d-8f4d-554e-a23e-59b1ae60b75b100% [##################################################] Parts: 1/1, Checksum: 100%, Write/sec: 1.3M/s, Read/sec: 0B/sFinalizing...Total execution time: 3.139 sTotal bytes read : 0Total bytes written : 17,246Upload completed
Publishing the analysis
The final step in data submission is to set the state of the analysis to PUBLISHED. A published analysis signals Maestro to index the data making it available on the front end portal interface.
Publish Command
docker exec song-client sh -c "sing publish -a {AnalysisId}"
Expected Response
{"message":"AnalysisId 4d9ed1c5-1053-4377-9ed1-c51053f3771f successfully published"}
You should now be able to find you uploaded data on the front-end portal found at localhost:3000/explorer
 Help us make our guides better
Help us make our guides better
If you can't find what you're looking for please reach out to us on our Slack channel linked on the top right of your screen or by email at contact@overture.bio